Kaggle: Personalized Medicine: Redefining Cancer Treatment
Problem statement
Classify the given genetic variations/mutations based on evidence from text-based clinical literature.
Data sources
Get the data from Kaggle.
We have two data files: one contains the information about the genetic mutations and the other contains the clinical evidence (text) that human experts/pathologists use to classify the genetic mutations. Both these data files have a common column called ID.
Data file’s information:
- training_variants (ID , Gene, Variations, Class)
- training_text (ID, Text)
Mapping to a ML problem
There are 9 distint classes of mutations to classify into, hence a multi-class classfication problem with metric as log-loss.
Exploratory Data Analysis
How data is presented?
First file
Number of data points : 3321
Number of features : 4
Features : [‘ID’ ‘Gene’ ‘Variation’ ‘Class’]
Second file
Number of data points : 3321
Number of features : 2
Features : [‘ID’ ‘TEXT’]
Merging both the files together based on ID. Next, we look for missing data which is present in TEXT attribute for 5 instances. Filling the NaN by combining Gene and Variation separated by a space.
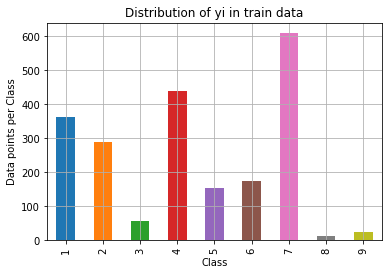
Proceeding with train(64%), test(20%) and validation(16%) split of the data. Since it is a multi class problem, we always split data following stratified sampling such that the distribution can be maintained equally. The distribution of classes in each data is shown in following graphs -


Featurizing Gene
There are 229 different categories of genes in the train data, and they are distibuted as follows
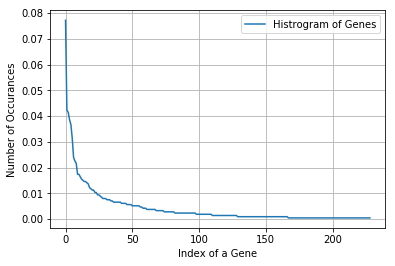
Next, we featurize Gene using one-hot encoding giving us 229 new features.
In test data 643 out of 665 data points are covered: 96.69%
In cross validation data 514 out of 532 data points are covered: 96.61%
Featurizing Variations
There are 1924 different categories of variations in the train data, and they are distibuted as follows

Next, we featurize Variation using one-hot encoding giving us 1924 new features.
In test data 64 out of 665 data points are covered: 9.62%
In cross validation data 56 out of 532 data points are covered: 10.52%
Featurizing Text
Total number of unique words in train data : 54850
Next, we featurize Text using one-hot encoding giving us 54850 new features.
97.125 % of word of test data appeared in train data
98.056 % of word of Cross Validation appeared in train data
=============================
One hot encoding features :
(number of data points * number of features) in train data = (2124, 57039)
(number of data points * number of features) in test data = (665, 57039)
(number of data points * number of features) in cross validation data = (532, 57039)
Machine Learning model
Applying balanced class linear SVM on one-hot encoded features gave us the following results -
For values of best alpha = 0.01 The train log loss is: 0.76
For values of best alpha = 0.01 The cross validation log loss is: 1.24
For values of best alpha = 0.01 The test log loss is: 1.15




